-
Introduction
-
Data collection and annotaion
-
Data statistics
-
User's guide
- Browse the data
- Search the data
- Detail of the data
-
Data download and feedback
Introduction
Radiotherapy is a commonly used therapeutic modality in cancer treatment, either alone or in combination with other treatment regimens in the clinic (Begg et al., 2011; Delaney et al., 2005). This treatment takes advantage of high intensity ionizing radiation to suppress tumour proliferation with no depth restriction and exerts its therapeutic effect mainly via DNA double-strand damage. The optimal goal of radiotherapy is to enhance the tumour radiation response specifically and to reduce toxicity in the surrounding normal tissue (Pavlopoulou et al., 2017). However, patients receiving radiotherapy may experience adverse treatment effects due to radioresistance and undifferentiated radiation that is toxic to normal tissue. Acute or early toxicity in normal tissue usually occurs during or within weeks following the treatment. Therefore, developing a precise treatment with appreciable radiosensitivity capacity for cancer tissues is an attractive strategy, which can overcome the undesirable side-effects that impede the use of radiotherapy (Bernier, 2017; Chen and Kuo, 2017).
Establishing effective treatment methods should be based on comprehensive understanding of the radiosensitivity mechanisms. There has been sustained interest in assessing the biological effects of radiosensitivity due to its potential clinical value (Ni et al., 2017; Yong et al., 2017). Generally, radiosensitivity studies have focused on the molecular mechanisms of radiotherapy with the goal to discover the regulation factors (compounds, genes or gene products) that can enhance cancer radiosensitivity (Goto et al., 2017; Pavlopoulou et al., 2017). It has been confirmed that DNA repair efficiency, cell cycle arrest, apoptosis and autophagy are the main mechanisms associated with radiosensitivity in many cancers (Forker et al., 2015; Gerweck and Wakimoto, 2016; Ni et al., 2017; Xin et al., 2017). During the past decades, many radiotherapy regulation factors have been identified. For example, HIF1A (hypoxia inducible factor 1 alpha subunit) knockouts enhance radiosensitivity by suppressing the DNA double-strand break (DSB) repair pathway (Elser et al., 2008). Similarly, a decrease in the long non-coding RNA HOTAIR (HOX transcript antisense RNA) efficiently enhances radiosensitivity in breast cancer via the Akt pathway (Zhou et al., 2017). Overexpression of hsa-mir-503 reinforces radiation responses in laryngeal carcinoma (Ma et al., 2017). Finally, curcumin has been shown to enhance the radiosensitivity of renal cancer cells by suppressing the NF-κB signalling pathway (Li et al., 2017).
To our knowledge, there is currently no database that specifically focuses on the regulation factors of radiotherapy, although some literature reviews have already summarized several specific mechanisms and listed certain radiosensitivity factors (Pavlopoulou et al., 2017). Undigested data buried in the extensive radiotherapy literature may limit the ability to develop a comprehensive understanding of radiosensitivity and impede improvements in radiotherapy. To fill this information gap, we developed a literature-based radiosensitivity regulation factor database (dbCRSR), which is proposed to assist in the investigation of the detailed mechanisms involved in radiotherapy efficacy and improve clinical treatment.
Data collection and annotation
First, to obtain the relevant publications, we performed an extensive PubMed literature query on October 9, 2017, using a list of keywords. After identifying the appropriate literature, we read through each paper to collect the pertinent information, including publication information, cancer type, radiation ray type, and radiosensitivity regulation factors (e.g., genes, miRNAs, lncRNAs, and compounds) along with the regulation mode of each factor. Labels “up” or “down” were used to indicate up-regulation or down-regulation of the regulation factors that enhance radiosensitivity. Second, multiple online tools were used to annotate the raw data from PubMed. To enrich the data and to facilitate users, we extracted candidate miRNAs and drugs from the miRTarBase and DrugBank databases, respectively, according to the relevant gene information.
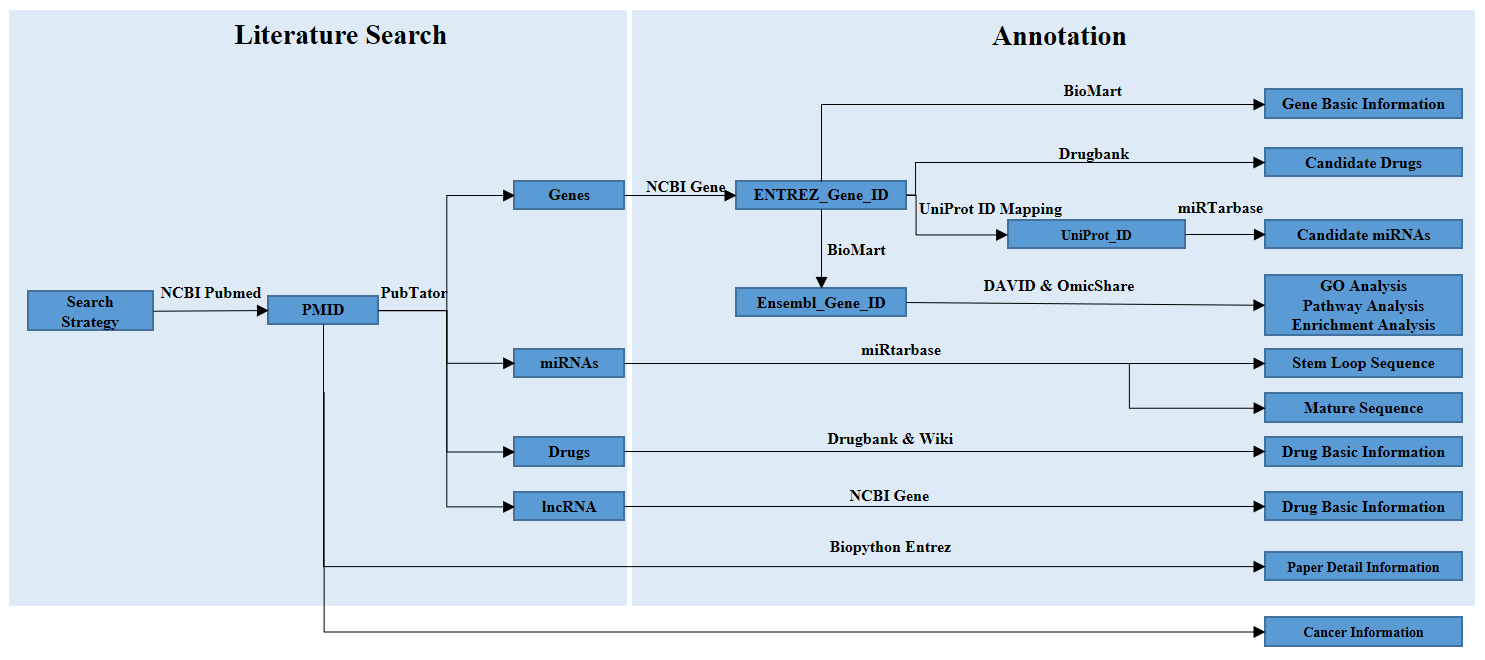
Data statistics
According to the data we have collected, statistic result can be found blow.
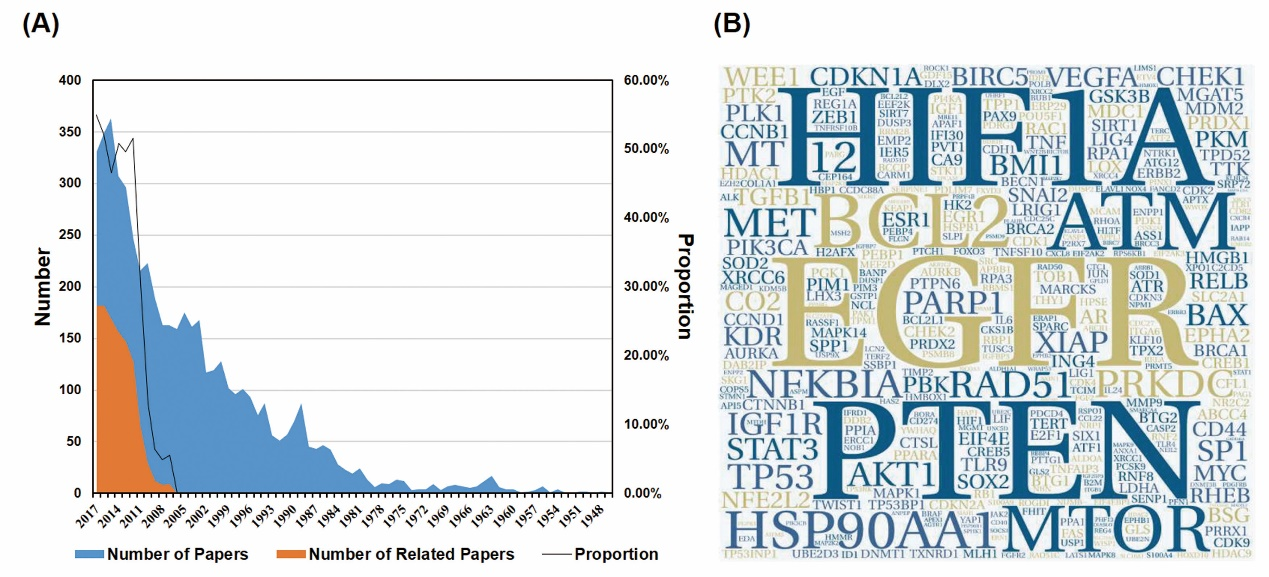
An overview of the data we collected. (A) The blue color indicates the number of the paper we gathered from PubMed. The Orange color indicates the number of the paper related to the research topic. The black line indicates the proportion (orange/blue) of each year, reflecting the percentage of the effective papers. (B) The hotspot genes of this area. The larger word size means the more frequency of the genes. This format is useful for quickly perceiving the most prominent terms.
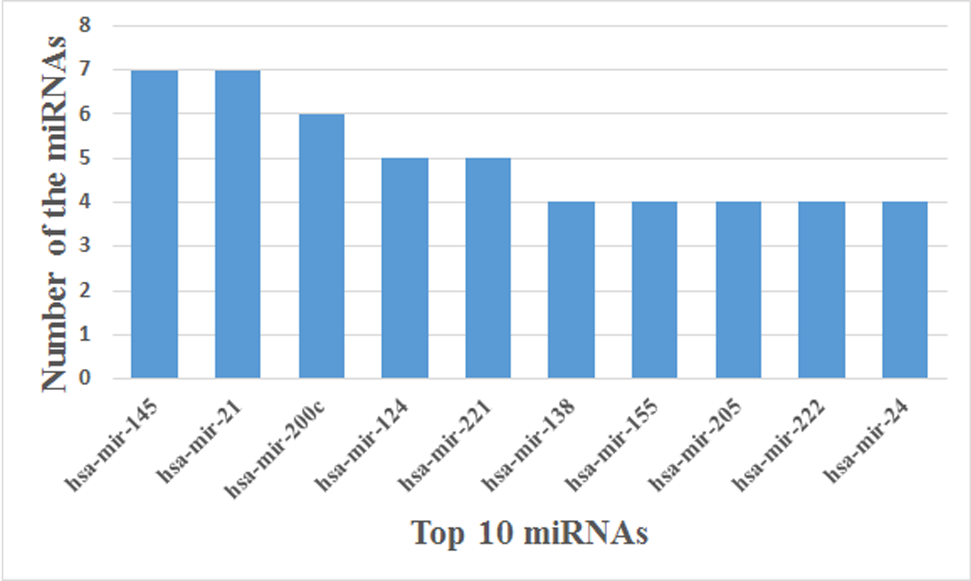
10 highest frequency miRNAs.
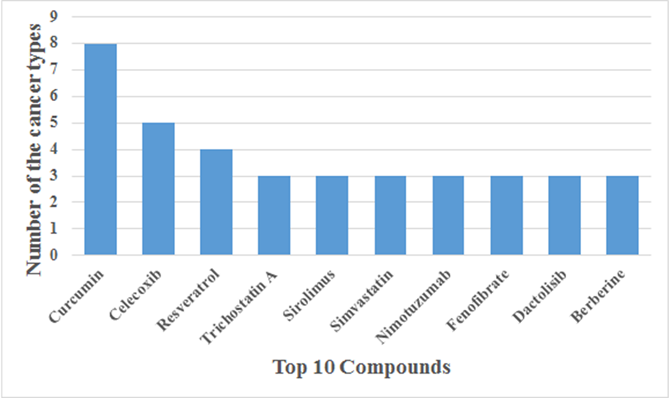
Top 10 compounds function in multiple cancers.
User's guide
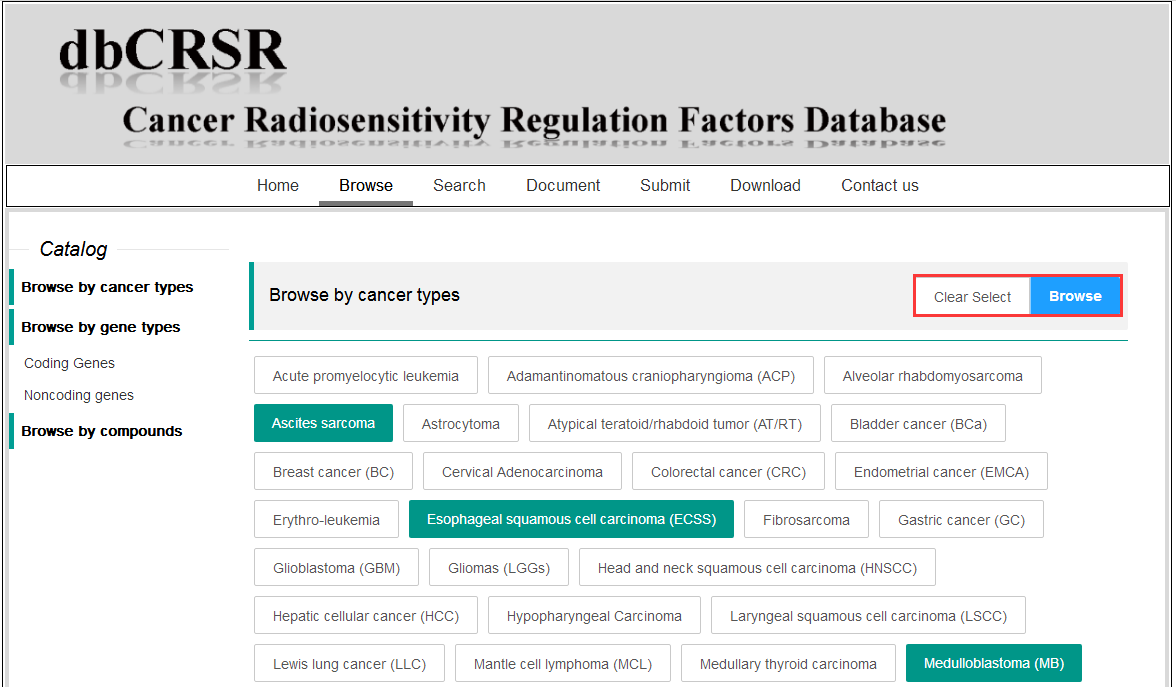
1. Browse the data
To browse the data, multiple cancer terms can be selected simultaneously.
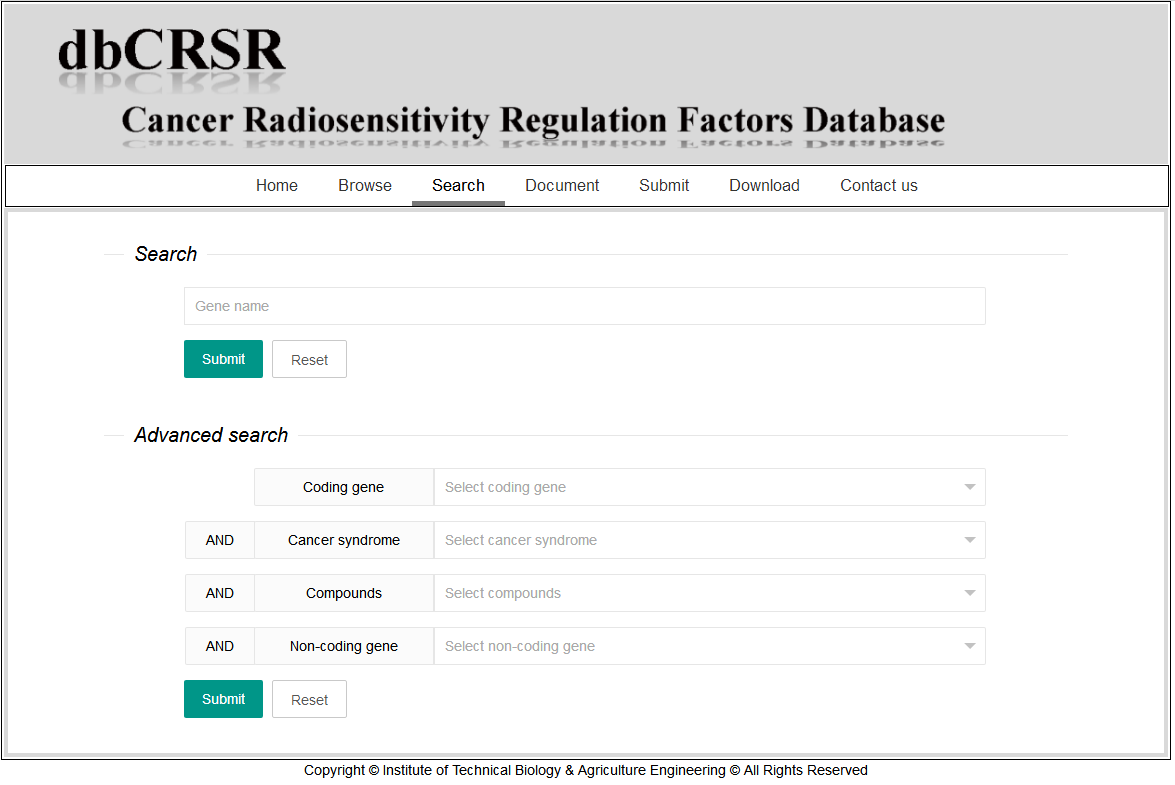
2. Search the data
Both basic and advanced strategy are available in our website.
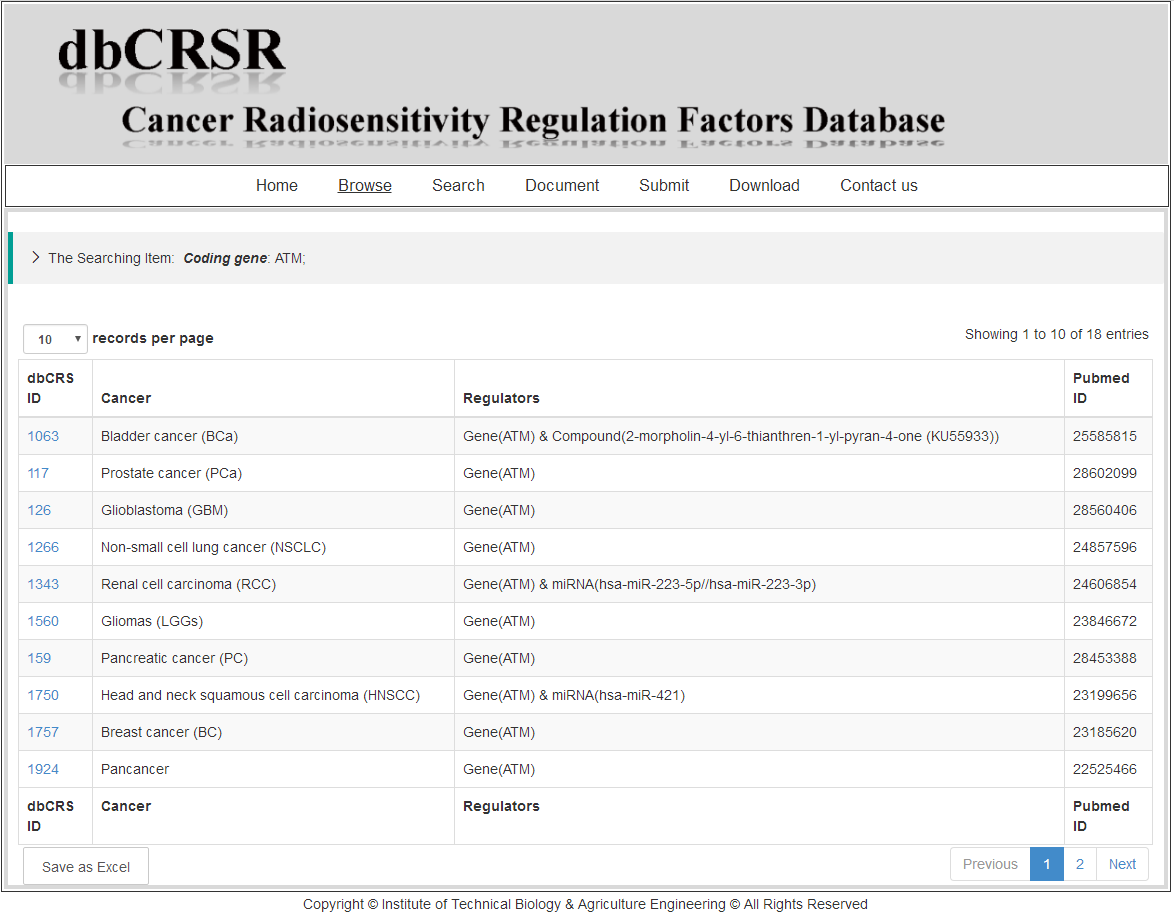
Take for example, after searching the gene "ATM", the record page will be returned. The first column is the internal ID we determined. The second column is the cancer type information retrieved from the original paper. The third column contains the regulators that could regulate radiosensitivity (each regulator separate from "&"). The last column is the PubMed ID information. Furthermore, users can turn to the detail information page by clicking the ID in the first column.
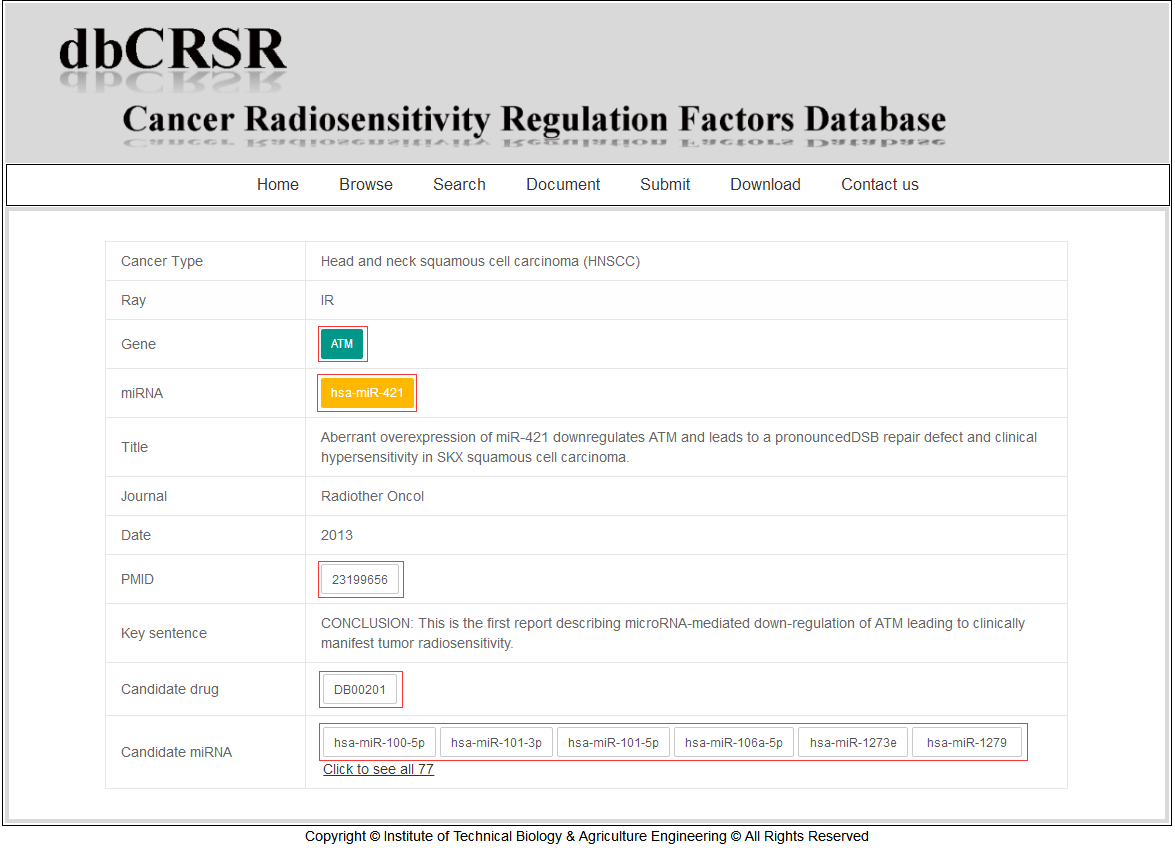
3. Detail of the data
The "orange" and "green" color are used to indicate up-regulation and down-regulation of the regulation factors that enhance radiosensitivity, respectively. The "candidate drug" and "candidate miRNA" imply that they have potential function in regulation of radiosensitivity-related genes. These information was annotated from the miRTarBase and DrugBank databases. For detailed information, please refer to the links we provided in the box.
Data download and feedback
You can freely download our data for academic researchers, but not for profit purposes.
We also hope you can help us to improve our database. If you have any suggestion to add a new comment to records in current dbCRSR or to revise wrong information in current dbCRSR, please send us email gpz@ipp.ac.cn directly.