-
About dbCID
-
Web Browser Requirements
-
Evidence System
-
Usage
- 1. Browse
- 2. Search
- 3. Download
- 4. Contact
-
Database Summary
-
Version
About dbCID
While recent advances in next generation sequencing technologies have enabled the creation of a multitude of databases in cancer genomic, there is no comprehensive database focusing on the annotation of driver indel yet. Therefore, we created the dbCID which is a collection of known indels that likely to be engaged in cancer development, progression or therapy. It currently contains experimentally supported and putative driver indels derived from manual curation of literature. For each indel, we have curated the position information (genomic, coding DNA, and protein levels), specific diseases, drug sensitivity information (partial) as well as evidence sentences. Evidence information is classified using the levels and rules of Evidence System. The database can be used to improve the training of prediction algorithms and evaluate the methods for predicting the effects of variations.
To obtain genomic positions, we used TransVar (http://bioinformatics.mdanderson.org/transvarweb/) through entering genes and their associated cDNA changes and mapped them to the results of the longest possible transcripts. Genomic positions that failed to match the canonical cDNA at the specified site were substituted by a dot (.). To acquire standard disease terminology,
we mapped the related disease onto DOIDs (Disease Ontology IDs, http://www.disease-ontology.org/).
Please cite the paper, if you are using the information in the database:
Yue Z, Zhao L, Cheng N, et al. dbCID: a manually curated resource for exploring the driver indels in human cancer. Briefings in Bioinformatics, Volume 20, Issue 5, September 2019, Pages 1925–1933
Datasets used in this article:
Download the training dataset for developing the prediction algorithm.
Download the datasets used in Figure 3.
Web Browser Requirements
The dbCID requires a modern web browser with JavaScript and cookies enabled. To view the complex details, pop-ups must not be blocked. The following browsers have been thoroughly tested with dbCID:
- Mozilla Firefox, version 4 or above
- Internet Explorer, versions 9 or above
- Chrome, version 5 or above
Evidence System
Rules for indel Entry into dbCID.
| Rule No. | Details |
|---|---|
| Rule 1 | Induced development, recurrence or metastasis of cancer. |
| Rule 2 | Associated with increased sensitivity or resistance to a drug. |
| Rule 3 | Induced change of function of gene product significantly. |
| Rule 4 | Had a higher recurrence frequency in cancer patients compared to the case of healthy controls. |
| Rule 5 | Located in an important region in gene or protein, such as a binding or catalytic site. |
Levels of evidence for indels in dbCID.
| Level No. | Details |
|---|---|
| Level 1 (in vivo) |
Indel is regarded as a driver based on evidence from functional experiments in vivo. |
| Level 2 (In vitro) |
Indel is regarded as a driver based on evidence from functional experiments In vitro. |
| Level 3 (Putative) |
Indel is a putative driver based on evidence such as a high recurrence frequency in cancer patients, an important location of protein and so on. |
Usage
In dbCID, we tried to make it powerful and convenient to be used. This Usage is prepared for the online service. The dbCID provides the browse function, search function and download function at present.
Capitalised titles correspond to column headings in the web page tables:
- GENE: The official gene symbol
- DISEASE: disease terminology in Disease Ontology
- DOID: Disease Ontology identifier
- Type: deletion, insertion, duplication or complex (insertion occurs simultaneously with deletion)
- Effect: frameshift or inframe
- Drug: sensitive or resistant to a certain drug
- HIGHEST LEVEL: The highest evidence level of indels across specified both disease and gene
1. Browse
You can select one or more of the four options listed in the browse area (Diease, Gene, Indel and Evidence). The Indel option only can be available after a gene is selected.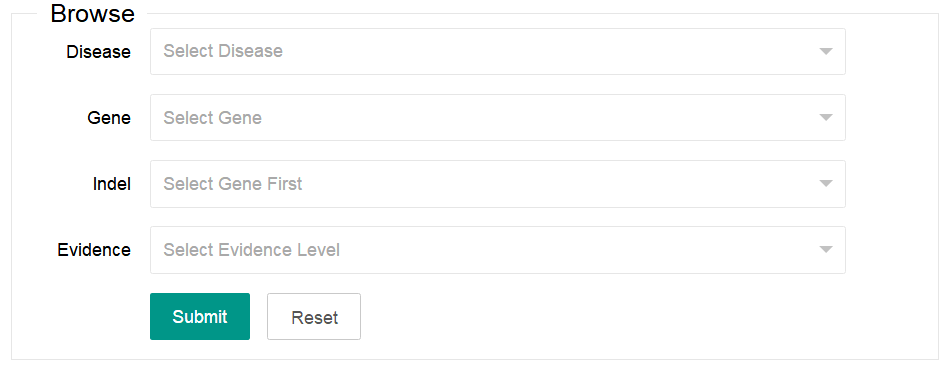
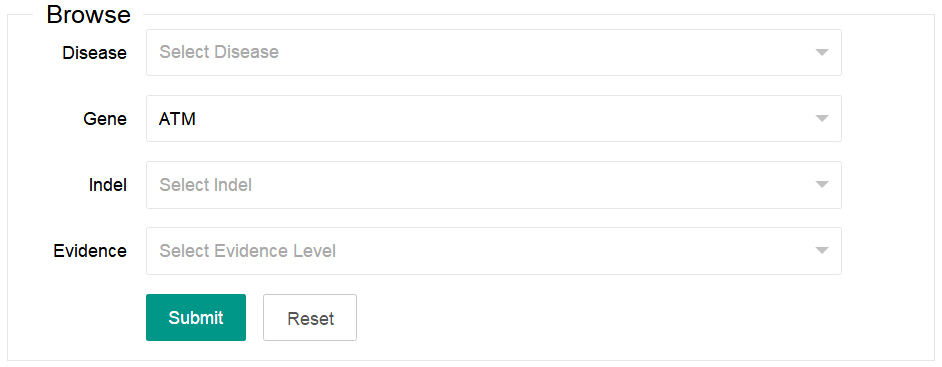
2. Search
You can input one keyword to search the dbCID. The search fields include disease name, gene name or indel (gDNA, cDNA or protein sequences). Only the GRCh38 genome assembly is supported.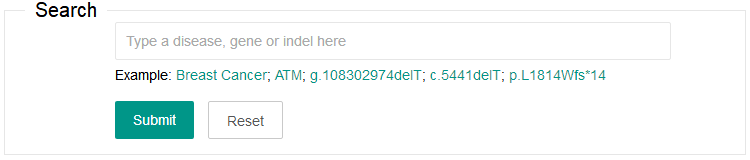
3. Download
We provide the option to download the full database. If you'd like to download it, please click to download the data for each level.
4. Contact
I have a few questions which are not listed above, how can I contact the authors of dbCID?Please contact Dr. Junfeng Xia (Email: jfxia@ahu.edu.cn) for details.
Database Summary (current version)
(A) Database statistics
(B) Type distribution of unique indels
| Gene | Indel | Disease | PMID | Entry | Level 1 (in vivo) |
Level 2 (In vitro) |
Level 3 (putative) |
| 67 | 895 | 57 | 270 | 1569 | 68 | 196 | 1305 |
| Deletion | Insertion | Duplication | Complex | Total | |
| FS | 517 | 92 | 102 | 27 | 738 |
| IF | 88 | 28 | 10 | 31 | 157 |
| Total | 605 | 120 | 112 | 58 | 895 |
FS: frameshift; IF: inframe; Complex: insertion occurs simultaneously with deletion
Version
The current version number is 1.0 - January, 2018. The most recent update to data was on January 10th, 2018.