1. Input for the online version
You can input the frameshift indels together with the transcript stable IDs from Ensembl. The transcript stable IDs can be acquired through BioMart data mining tool and the cDNA information should conform to the human genome variation society (HGVS) standard. Transcript and cDNA information should be separated with the colon character ":".
2. Input for the locally running version
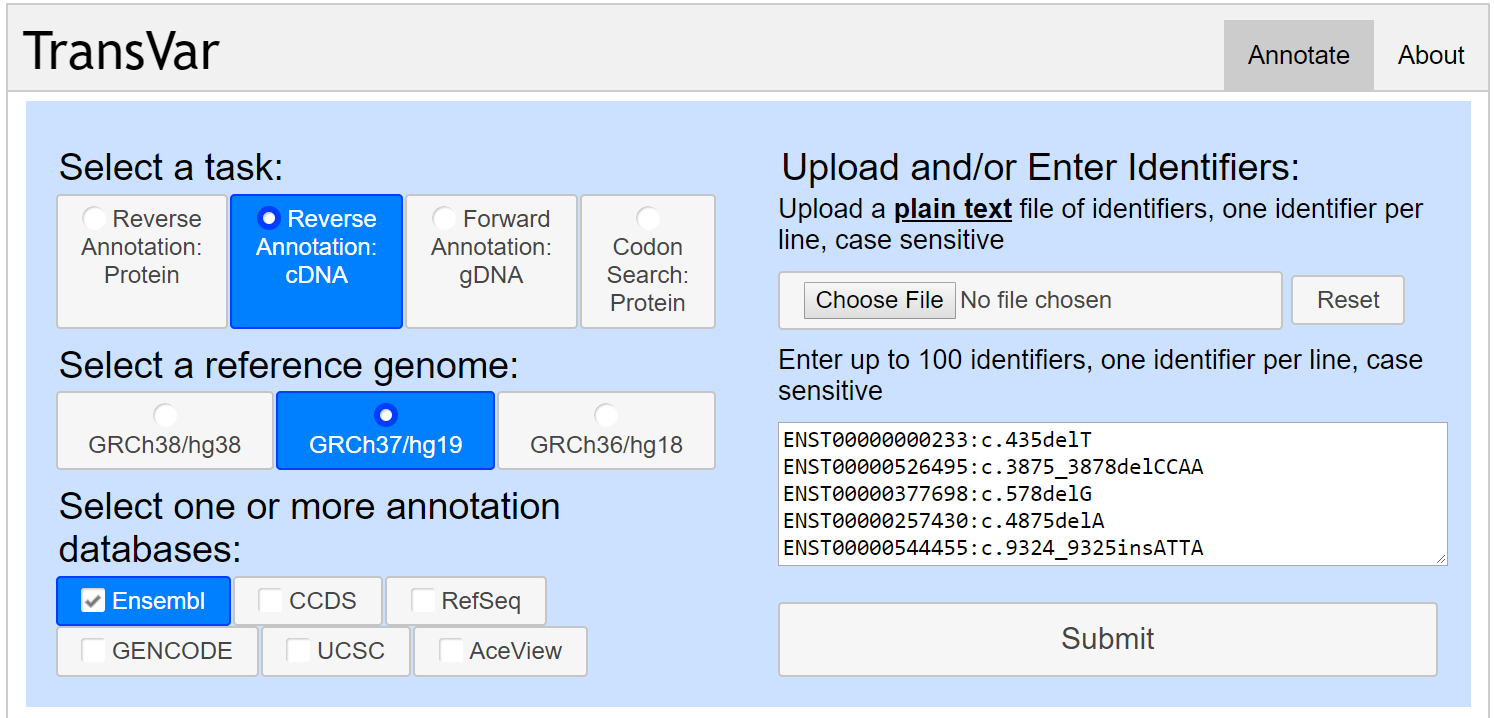
If you'd like to use the locally running version of PredCID, the inputs requires the position information of indels both in cDNA and genome, which can be annotated by the tool TransVar. For the selection of various options in TransVar, please see the figure below.

3. Quick start for the locally running version
The following shows the demo code of PredCID.
Please feel free to contact us if you need any help: zhenyuyue@ahau.edu.cn
setwd("/path/to/this/file/PredCID/")
library(bitops)
library(methods)
library(stringr)
library(biomaRt)
library(RCurl)
library(xgboost)
source("./scripts/getResults.R")
getResults("example","outputFileName")#example.csv is a file containing frameshift indels.
4. Outputs
In general, the predictions are done on the fly, and in seconds to minutes, depending mainly on the size of the input data. Users will receive an email attached with the result file. For each input, the model returns a score between zero and one, where indels with higher scores are more likely to be driver frameshift indel. Here, the indels with scores greater than 0.5 are predicted as drivers.
